The Singh Lab


WELCOME TO THE SINGH LAB
Cereals Epigenomics
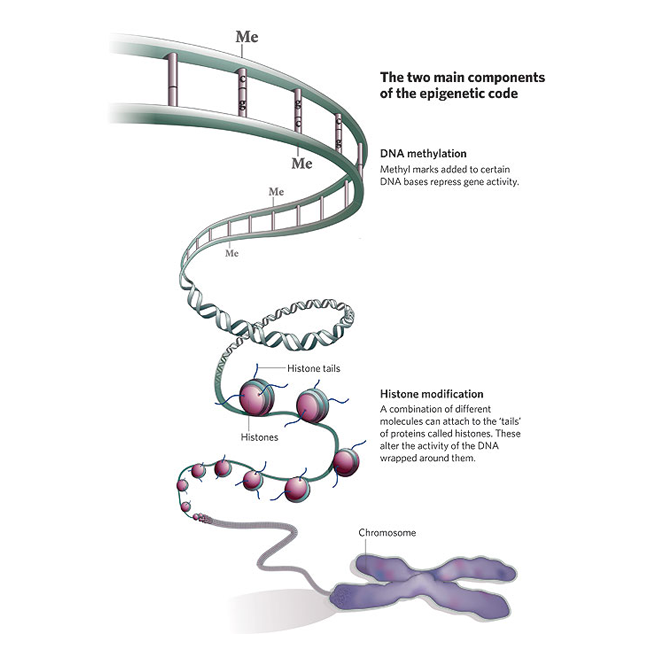
Epigenetic modifications provide important clues regarding gene regulation mechanisms. For example, RNA-directed DNA methylation (RdDM) pathway targets many biological processes including silencing of transposable element and genome stability, plant development and reproduction, short and long-range signaling, and stress response (Erdmann and Picard 2020, PLOS Genetics, 16 (10): e1009034). In our previous studies, association of the key gene of RdDM, ARGONAUTE4_9 with plant reproductive process and seed dormancy (Singh et al. 2013, PLOS One, 8(10): e77009) prompted us to elucidate the role of RdDM in agronomically important traits such as Pre-Harvest Sprouting in small grain cereals. We are also investigating the miR-SPL (microRNA-SQUAMOSA promoter-binding protein-Like) module for its involvement in the developmental processes in cereals, such as barley (Tripathi et al., 2018 Scientific Reports 8:7085) and Brachypodium (Tripathi et al., 2020 Scientific Reports 10; 15302) and currently is the key component of our epigenetics research.
Genetic transformation and gene editing in functional genomics
While the conventional breeding relied heavily on natural variations, the development of genetic transformation technologies provides an alternative approach in crop improvement. In the last few years, the emergence of gene editing technique became a major advance for site-directed mutagenesis first in animals, latter in plant species. We are one of the few labs worldwide working on cereal transformation and successfully developed oat and barley transformants tagged with Ac/Ds transposons, overexpressing gene of interests or down-regulating target genes. More recently, we are using CRISPR-Cas9 system to perform gene editing in oat and barley to study economically important traits.

High quality & nutritionally enhanced crops
We have been improving a variety of quality traits in cereal crops to meet the needs of different end users. For this purpose, we are exploring cysteine rich proteins (Thaumatin-Like Sweet Proteins), protein-carbohydrate interaction and redox regulation. We recently discovered a gene, HvTLP8 (thaumatin-like protein 8), redox-mediated controls the level of β-glucan in barley to enhance malt quality (Singh et al. 2017, PNAS, 114(29): 7725-7730). Meanwhile, we are targeting oil, starch, other nutritional traits in oat, barley, and field pea to add value to crop production.
More resources
Thank You
We would like to thank the following collaborators for their support.







